Aluminium »
PDB 2xzl-3wgu »
3cf1 »
Aluminium in PDB 3cf1: Structure of P97/Vcp in Complex with Adp/Adp.Alfx
Protein crystallography data
The structure of Structure of P97/Vcp in Complex with Adp/Adp.Alfx, PDB code: 3cf1
was solved by
J.M.Davies,
B.Delabarre,
A.T.Brunger,
W.I.Weis,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.87 / 4.40 |
| Space group | I 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 162.660, 178.020, 321.140, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 22.9 / 28.6 |
Other elements in 3cf1:
The structure of Structure of P97/Vcp in Complex with Adp/Adp.Alfx also contains other interesting chemical elements:
| Fluorine | (F) | 9 atoms |
Aluminium Binding Sites:
The binding sites of Aluminium atom in the Structure of P97/Vcp in Complex with Adp/Adp.Alfx
(pdb code 3cf1). This binding sites where shown within
5.0 Angstroms radius around Aluminium atom.
In total 3 binding sites of Aluminium where determined in the Structure of P97/Vcp in Complex with Adp/Adp.Alfx, PDB code: 3cf1:
Jump to Aluminium binding site number: 1; 2; 3;
In total 3 binding sites of Aluminium where determined in the Structure of P97/Vcp in Complex with Adp/Adp.Alfx, PDB code: 3cf1:
Jump to Aluminium binding site number: 1; 2; 3;
Aluminium binding site 1 out of 3 in 3cf1
Go back to
Aluminium binding site 1 out
of 3 in the Structure of P97/Vcp in Complex with Adp/Adp.Alfx
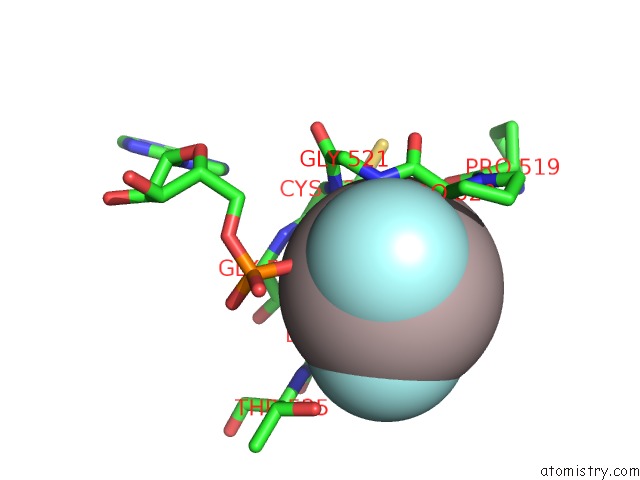
Mono view
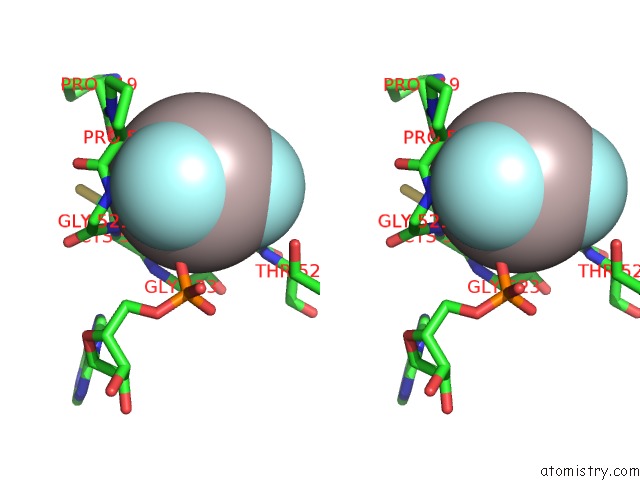
Stereo pair view

Mono view

Stereo pair view
A full contact list of Aluminium with other atoms in the Al binding
site number 1 of Structure of P97/Vcp in Complex with Adp/Adp.Alfx within 5.0Å range:
|
Aluminium binding site 2 out of 3 in 3cf1
Go back to
Aluminium binding site 2 out
of 3 in the Structure of P97/Vcp in Complex with Adp/Adp.Alfx
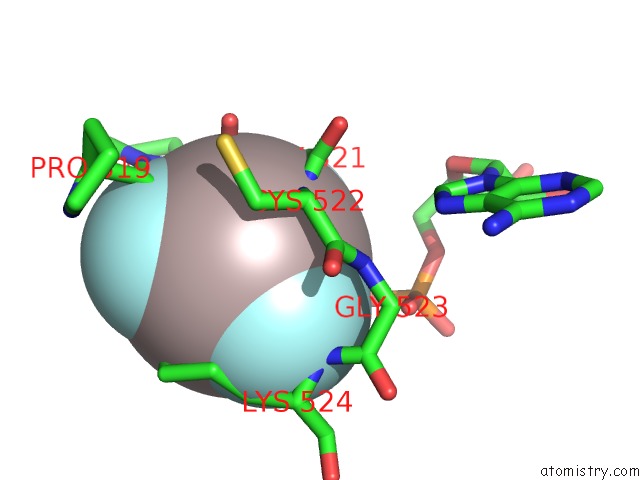
Mono view
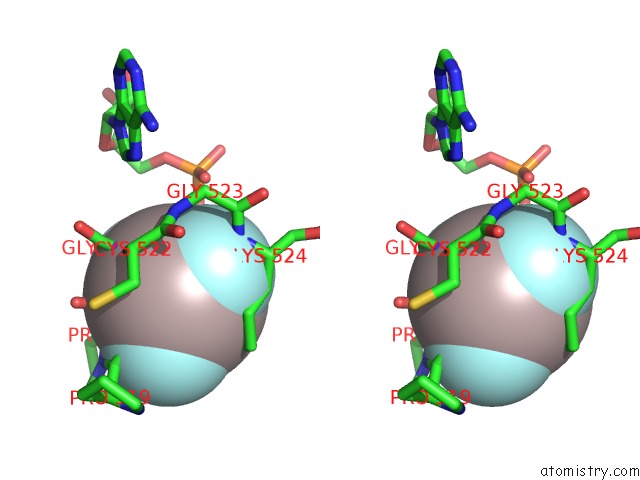
Stereo pair view

Mono view

Stereo pair view
A full contact list of Aluminium with other atoms in the Al binding
site number 2 of Structure of P97/Vcp in Complex with Adp/Adp.Alfx within 5.0Å range:
|
Aluminium binding site 3 out of 3 in 3cf1
Go back to
Aluminium binding site 3 out
of 3 in the Structure of P97/Vcp in Complex with Adp/Adp.Alfx
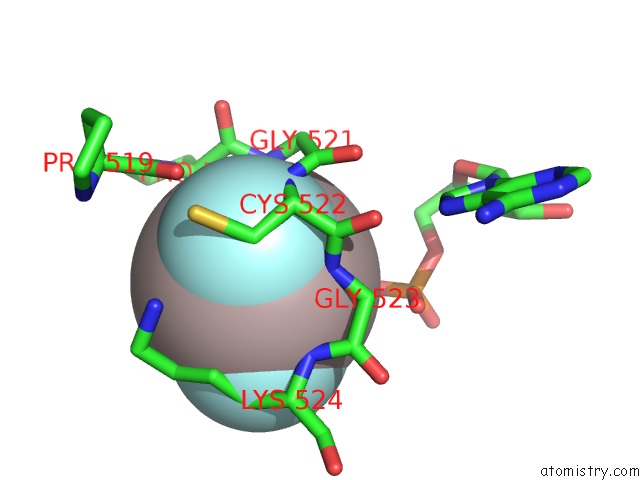
Mono view
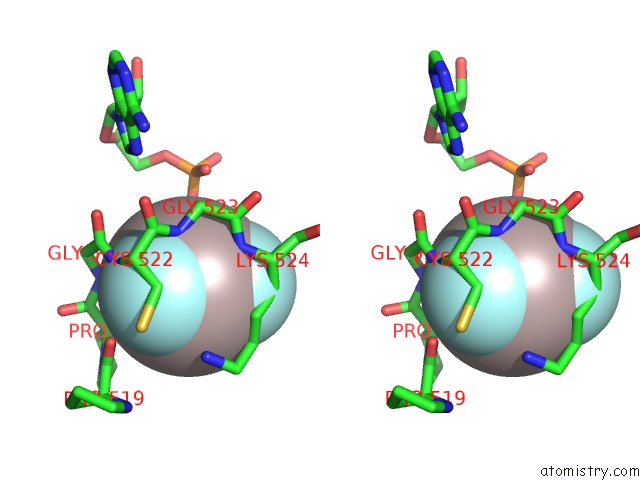
Stereo pair view

Mono view

Stereo pair view
A full contact list of Aluminium with other atoms in the Al binding
site number 3 of Structure of P97/Vcp in Complex with Adp/Adp.Alfx within 5.0Å range:
|
Reference:
J.M.Davies,
A.T.Brunger,
W.I.Weis.
Improved Structures of Full-Length P97, An Aaa Atpase: Implications For Mechanisms of Nucleotide-Dependent Conformational Change. Structure V. 16 715 2008.
ISSN: ISSN 0969-2126
PubMed: 18462676
DOI: 10.1016/J.STR.2008.02.010
Page generated: Sun Jul 6 21:45:06 2025
ISSN: ISSN 0969-2126
PubMed: 18462676
DOI: 10.1016/J.STR.2008.02.010
Last articles
F in 7N72F in 7N70
F in 7N3J
F in 7N6F
F in 7N66
F in 7N33
F in 7N4Q
F in 7N4N
F in 7N2A
F in 7MXY