Aluminium »
PDB 2xzl-3wgu »
3d7m »
Aluminium in PDB 3d7m: Crystal Structure of the G Protein Fast-Exchange Double Mutant I56C/Q333C
Protein crystallography data
The structure of Crystal Structure of the G Protein Fast-Exchange Double Mutant I56C/Q333C, PDB code: 3d7m
was solved by
M.A.Funk,
A.M.Preininger,
W.M.Oldham,
S.M.Meier,
H.E.Hamm,
T.M.Iverson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.90 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 79.659, 79.659, 114.587, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 24.9 / 29.4 |
Other elements in 3d7m:
The structure of Crystal Structure of the G Protein Fast-Exchange Double Mutant I56C/Q333C also contains other interesting chemical elements:
| Fluorine | (F) | 4 atoms |
| Magnesium | (Mg) | 1 atom |
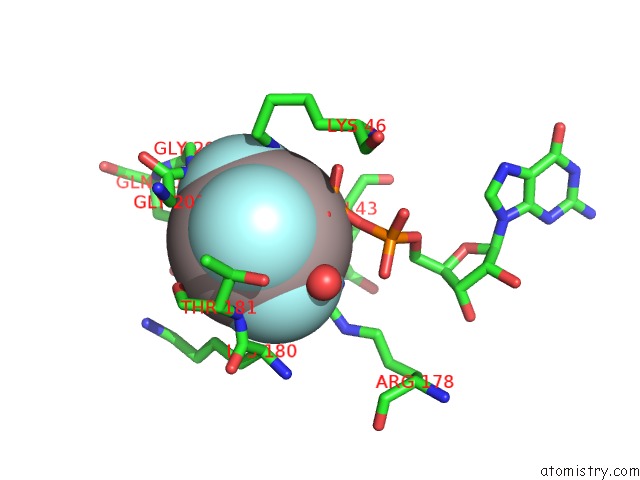
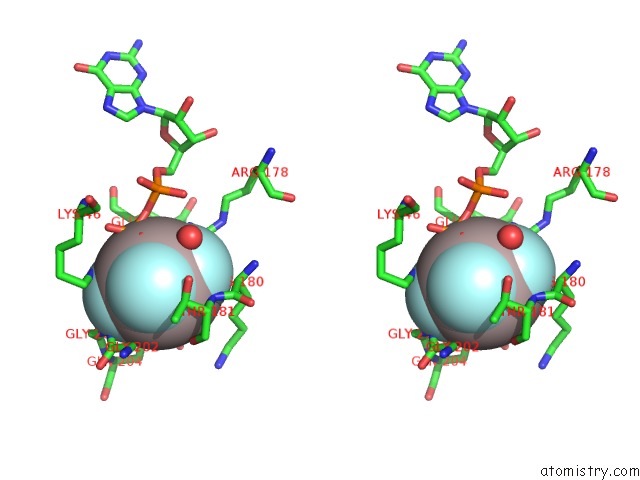
Aluminium Binding Sites:
The binding sites of Aluminium atom in the Crystal Structure of the G Protein Fast-Exchange Double Mutant I56C/Q333C
(pdb code 3d7m). This binding sites where shown within
5.0 Angstroms radius around Aluminium atom.
In total only one binding site of Aluminium was determined in the Crystal Structure of the G Protein Fast-Exchange Double Mutant I56C/Q333C, PDB code: 3d7m:
In total only one binding site of Aluminium was determined in the Crystal Structure of the G Protein Fast-Exchange Double Mutant I56C/Q333C, PDB code: 3d7m:
Aluminium binding site 1 out of 1 in 3d7m
Go back to
Aluminium binding site 1 out
of 1 in the Crystal Structure of the G Protein Fast-Exchange Double Mutant I56C/Q333C
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Aluminium with other atoms in the Al binding
site number 1 of Crystal Structure of the G Protein Fast-Exchange Double Mutant I56C/Q333C within 5.0Å range:
|
Reference:
A.M.Preininger,
M.A.Funk,
W.M.Oldham,
S.M.Meier,
C.A.Johnston,
S.Adhikary,
A.J.Kimple,
D.P.Siderovski,
H.E.Hamm,
T.M.Iverson.
Helix Dipole Movement and Conformational Variability Contribute to Allosteric Gdp Release in Galphai Subunits. Biochemistry V. 48 2630 2009.
ISSN: ISSN 0006-2960
PubMed: 19222191
DOI: 10.1021/BI801853A
Page generated: Sun Jul 6 21:47:22 2025
ISSN: ISSN 0006-2960
PubMed: 19222191
DOI: 10.1021/BI801853A
Last articles
Cl in 7ZSGCl in 7ZNM
Cl in 7ZSF
Cl in 7ZQX
Cl in 7ZR4
Cl in 7ZR3
Cl in 7ZQQ
Cl in 7ZQI
Cl in 7ZPG
Cl in 7ZL7