Aluminium »
PDB 2xzl-3wgu »
3i62 »
Aluminium in PDB 3i62: Structure of MSS116P Bound to Ssrna and Adp-Aluminum Fluoride
Protein crystallography data
The structure of Structure of MSS116P Bound to Ssrna and Adp-Aluminum Fluoride, PDB code: 3i62
was solved by
M.Del Campo,
A.M.Lambowitz,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 33.57 / 1.95 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 88.826, 126.111, 55.770, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.8 / 20.9 |
Other elements in 3i62:
The structure of Structure of MSS116P Bound to Ssrna and Adp-Aluminum Fluoride also contains other interesting chemical elements:
| Fluorine | (F) | 4 atoms |
| Magnesium | (Mg) | 1 atom |
Aluminium Binding Sites:
The binding sites of Aluminium atom in the Structure of MSS116P Bound to Ssrna and Adp-Aluminum Fluoride
(pdb code 3i62). This binding sites where shown within
5.0 Angstroms radius around Aluminium atom.
In total only one binding site of Aluminium was determined in the Structure of MSS116P Bound to Ssrna and Adp-Aluminum Fluoride, PDB code: 3i62:
In total only one binding site of Aluminium was determined in the Structure of MSS116P Bound to Ssrna and Adp-Aluminum Fluoride, PDB code: 3i62:
Aluminium binding site 1 out of 1 in 3i62
Go back to
Aluminium binding site 1 out
of 1 in the Structure of MSS116P Bound to Ssrna and Adp-Aluminum Fluoride
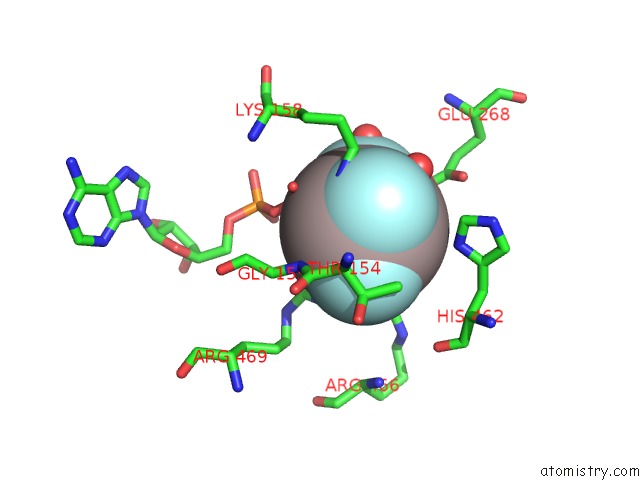
Mono view
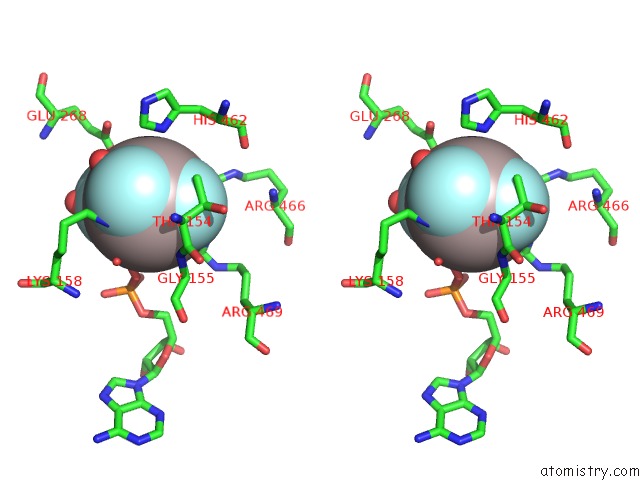
Stereo pair view

Mono view

Stereo pair view
A full contact list of Aluminium with other atoms in the Al binding
site number 1 of Structure of MSS116P Bound to Ssrna and Adp-Aluminum Fluoride within 5.0Å range:
|
Reference:
M.Del Campo,
A.M.Lambowitz.
Structure of the Yeast Dead Box Protein MSS116P Reveals Two Wedges That Crimp Rna Mol.Cell V. 35 598 2009.
ISSN: ISSN 1097-2765
PubMed: 19748356
DOI: 10.1016/J.MOLCEL.2009.07.032
Page generated: Sun Jul 6 21:48:15 2025
ISSN: ISSN 1097-2765
PubMed: 19748356
DOI: 10.1016/J.MOLCEL.2009.07.032
Last articles
Cl in 5X2OCl in 5X2N
Cl in 5X2M
Cl in 5X1W
Cl in 5X2K
Cl in 5X2I
Cl in 5X2C
Cl in 5X2A
Cl in 5X1V
Cl in 5X28