Aluminium »
PDB 3wgv-5c2j »
4m8n »
Aluminium in PDB 4m8n: Crystal Structure of PLEXINC1/RAP1B Complex
Protein crystallography data
The structure of Crystal Structure of PLEXINC1/RAP1B Complex, PDB code: 4m8n
was solved by
H.G.Pascoe,
Y.Wang,
C.A.Brautigam,
H.He,
X.Zhang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 41.05 / 3.29 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 76.276, 84.734, 138.752, 91.09, 95.15, 90.32 |
| R / Rfree (%) | 24.3 / 29.9 |
Other elements in 4m8n:
The structure of Crystal Structure of PLEXINC1/RAP1B Complex also contains other interesting chemical elements:
| Fluorine | (F) | 12 atoms |
| Magnesium | (Mg) | 4 atoms |
Aluminium Binding Sites:
The binding sites of Aluminium atom in the Crystal Structure of PLEXINC1/RAP1B Complex
(pdb code 4m8n). This binding sites where shown within
5.0 Angstroms radius around Aluminium atom.
In total 4 binding sites of Aluminium where determined in the Crystal Structure of PLEXINC1/RAP1B Complex, PDB code: 4m8n:
Jump to Aluminium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Aluminium where determined in the Crystal Structure of PLEXINC1/RAP1B Complex, PDB code: 4m8n:
Jump to Aluminium binding site number: 1; 2; 3; 4;
Aluminium binding site 1 out of 4 in 4m8n
Go back to
Aluminium binding site 1 out
of 4 in the Crystal Structure of PLEXINC1/RAP1B Complex
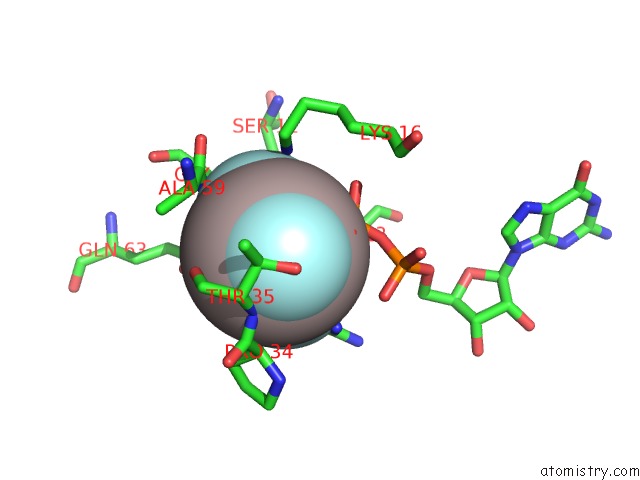
Mono view
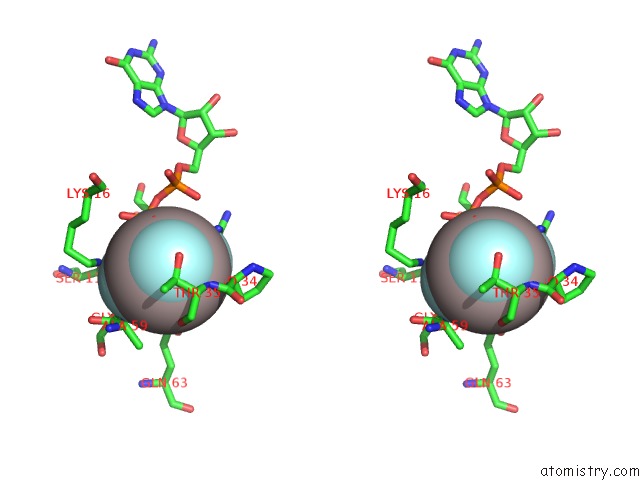
Stereo pair view

Mono view

Stereo pair view
A full contact list of Aluminium with other atoms in the Al binding
site number 1 of Crystal Structure of PLEXINC1/RAP1B Complex within 5.0Å range:
|
Aluminium binding site 2 out of 4 in 4m8n
Go back to
Aluminium binding site 2 out
of 4 in the Crystal Structure of PLEXINC1/RAP1B Complex
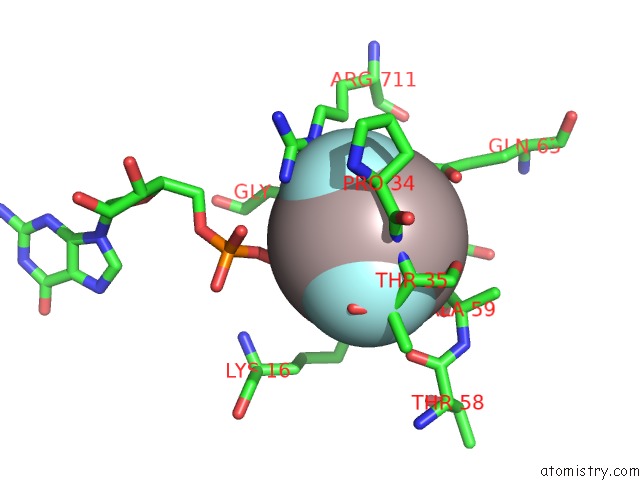
Mono view
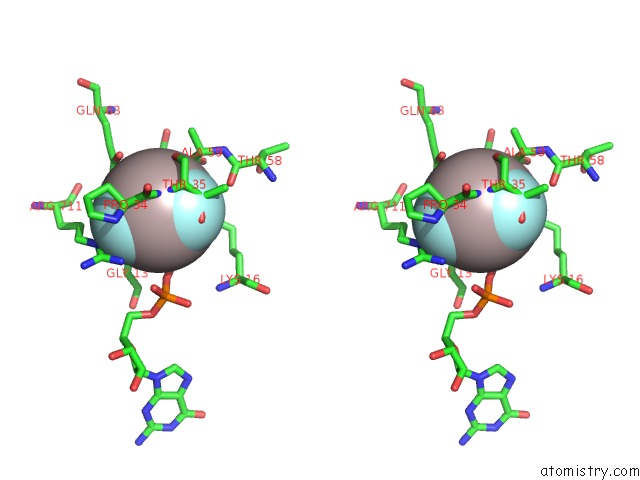
Stereo pair view

Mono view

Stereo pair view
A full contact list of Aluminium with other atoms in the Al binding
site number 2 of Crystal Structure of PLEXINC1/RAP1B Complex within 5.0Å range:
|
Aluminium binding site 3 out of 4 in 4m8n
Go back to
Aluminium binding site 3 out
of 4 in the Crystal Structure of PLEXINC1/RAP1B Complex
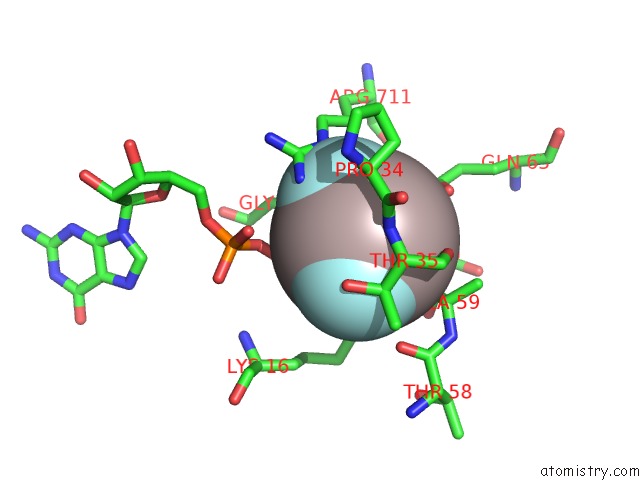
Mono view
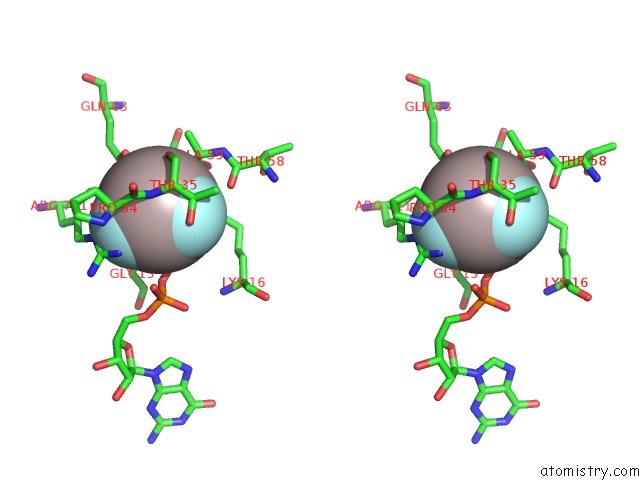
Stereo pair view

Mono view

Stereo pair view
A full contact list of Aluminium with other atoms in the Al binding
site number 3 of Crystal Structure of PLEXINC1/RAP1B Complex within 5.0Å range:
|
Aluminium binding site 4 out of 4 in 4m8n
Go back to
Aluminium binding site 4 out
of 4 in the Crystal Structure of PLEXINC1/RAP1B Complex
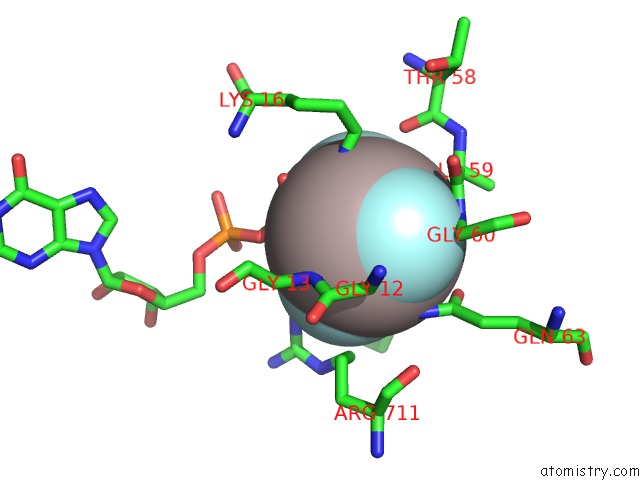
Mono view
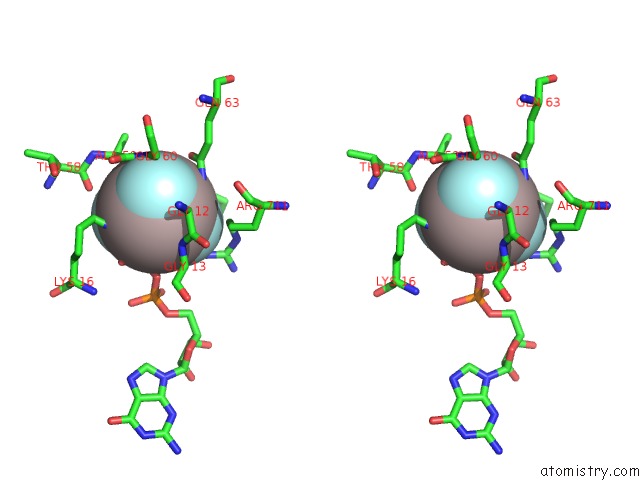
Stereo pair view

Mono view

Stereo pair view
A full contact list of Aluminium with other atoms in the Al binding
site number 4 of Crystal Structure of PLEXINC1/RAP1B Complex within 5.0Å range:
|
Reference:
Y.Wang,
H.G.Pascoe,
C.A.Brautigam,
H.He,
X.Zhang.
Structural Basis For Activation and Non-Canonical Catalysis of the Rap Gtpase Activating Protein Domain of Plexin. Elife V. 2 01279 2013.
ISSN: ESSN 2050-084X
PubMed: 24137545
DOI: 10.7554/ELIFE.01279
Page generated: Sun Jul 6 21:54:32 2025
ISSN: ESSN 2050-084X
PubMed: 24137545
DOI: 10.7554/ELIFE.01279
Last articles
Cl in 5PZWCl in 5PZV
Cl in 5PZR
Cl in 5PZS
Cl in 5PWC
Cl in 5PWD
Cl in 5PB6
Cl in 5PRC
Cl in 5PNY
Cl in 5POT