Aluminium »
PDB 5c2k-6i03 »
5o6e »
Aluminium in PDB 5o6e: Structure of SCPIF1 in Complex with Tttgggtt and Adp-ALF4
Enzymatic activity of Structure of SCPIF1 in Complex with Tttgggtt and Adp-ALF4
All present enzymatic activity of Structure of SCPIF1 in Complex with Tttgggtt and Adp-ALF4:
3.6.4.12;
3.6.4.12;
Protein crystallography data
The structure of Structure of SCPIF1 in Complex with Tttgggtt and Adp-ALF4, PDB code: 5o6e
was solved by
K.Y.Lu,
W.F.Chen,
S.Rety,
N.N.Liu,
X.G.Xu,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 65.21 / 3.35 |
| Space group | P 31 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 150.605, 150.605, 136.339, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 17.8 / 25.7 |
Other elements in 5o6e:
The structure of Structure of SCPIF1 in Complex with Tttgggtt and Adp-ALF4 also contains other interesting chemical elements:
| Fluorine | (F) | 8 atoms |
| Magnesium | (Mg) | 2 atoms |
| Chlorine | (Cl) | 1 atom |
Aluminium Binding Sites:
The binding sites of Aluminium atom in the Structure of SCPIF1 in Complex with Tttgggtt and Adp-ALF4
(pdb code 5o6e). This binding sites where shown within
5.0 Angstroms radius around Aluminium atom.
In total 2 binding sites of Aluminium where determined in the Structure of SCPIF1 in Complex with Tttgggtt and Adp-ALF4, PDB code: 5o6e:
Jump to Aluminium binding site number: 1; 2;
In total 2 binding sites of Aluminium where determined in the Structure of SCPIF1 in Complex with Tttgggtt and Adp-ALF4, PDB code: 5o6e:
Jump to Aluminium binding site number: 1; 2;
Aluminium binding site 1 out of 2 in 5o6e
Go back to
Aluminium binding site 1 out
of 2 in the Structure of SCPIF1 in Complex with Tttgggtt and Adp-ALF4
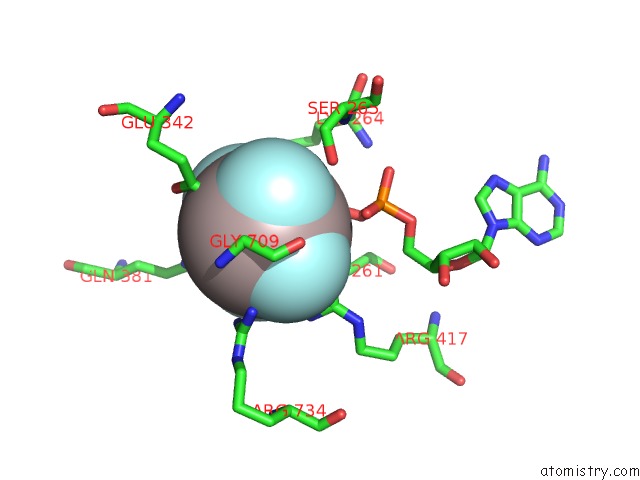
Mono view
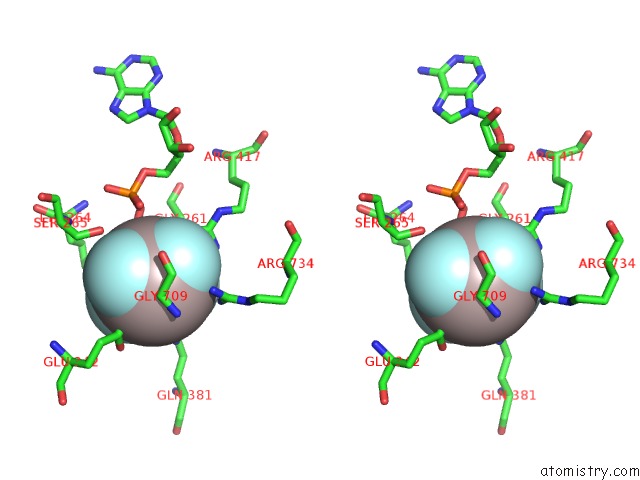
Stereo pair view

Mono view

Stereo pair view
A full contact list of Aluminium with other atoms in the Al binding
site number 1 of Structure of SCPIF1 in Complex with Tttgggtt and Adp-ALF4 within 5.0Å range:
|
Aluminium binding site 2 out of 2 in 5o6e
Go back to
Aluminium binding site 2 out
of 2 in the Structure of SCPIF1 in Complex with Tttgggtt and Adp-ALF4
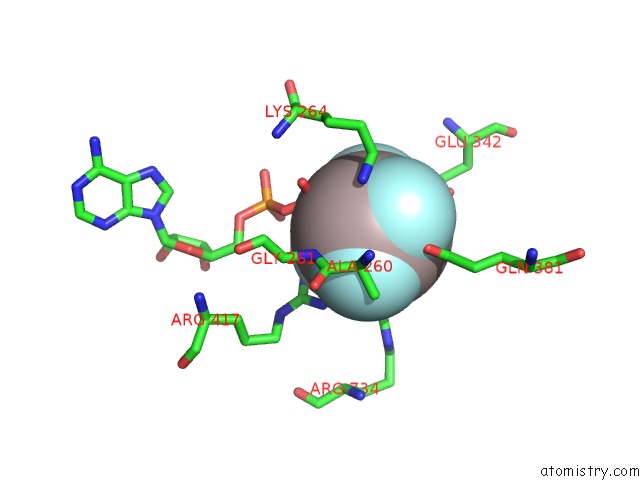
Mono view
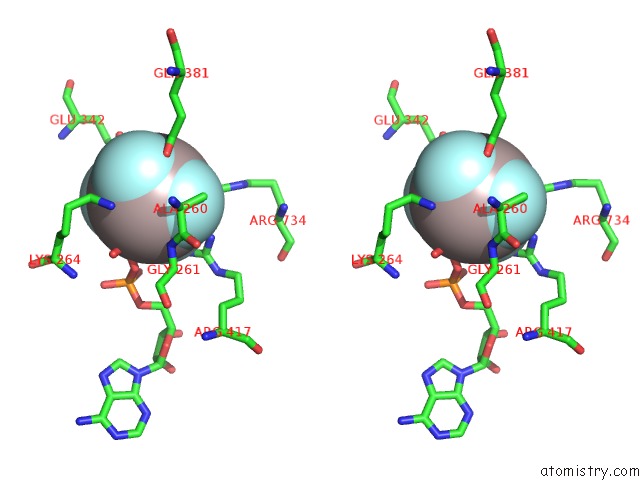
Stereo pair view

Mono view

Stereo pair view
A full contact list of Aluminium with other atoms in the Al binding
site number 2 of Structure of SCPIF1 in Complex with Tttgggtt and Adp-ALF4 within 5.0Å range:
|
Reference:
K.Y.Lu,
W.F.Chen,
S.Rety,
N.N.Liu,
W.Q.Wu,
Y.X.Dai,
D.Li,
H.Y.Ma,
S.X.Dou,
X.G.Xi.
Insights Into the Structural and Mechanistic Basis of Multifunctional S. Cerevisiae PIF1P Helicase. Nucleic Acids Res. V. 46 1486 2018.
ISSN: ESSN 1362-4962
PubMed: 29202194
DOI: 10.1093/NAR/GKX1217
Page generated: Sun Jul 6 21:57:25 2025
ISSN: ESSN 1362-4962
PubMed: 29202194
DOI: 10.1093/NAR/GKX1217
Last articles
Cl in 7VUECl in 7VUI
Cl in 7VUH
Cl in 7VUG
Cl in 7VU6
Cl in 7VS8
Cl in 7VTX
Cl in 7VTW
Cl in 7VTI
Cl in 7VTV