Aluminium »
PDB 5c2k-6i03 »
6b9f »
Aluminium in PDB 6b9f: Human ATL1 Mutant - F151S Bound to GDPALF4-
Protein crystallography data
The structure of Human ATL1 Mutant - F151S Bound to GDPALF4-, PDB code: 6b9f
was solved by
J.P.O'donnell,
H.Sondermann,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.68 / 1.90 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 49.613, 115.721, 184.300, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.8 / 22.2 |
Other elements in 6b9f:
The structure of Human ATL1 Mutant - F151S Bound to GDPALF4- also contains other interesting chemical elements:
| Fluorine | (F) | 8 atoms |
| Magnesium | (Mg) | 2 atoms |
Aluminium Binding Sites:
The binding sites of Aluminium atom in the Human ATL1 Mutant - F151S Bound to GDPALF4-
(pdb code 6b9f). This binding sites where shown within
5.0 Angstroms radius around Aluminium atom.
In total 2 binding sites of Aluminium where determined in the Human ATL1 Mutant - F151S Bound to GDPALF4-, PDB code: 6b9f:
Jump to Aluminium binding site number: 1; 2;
In total 2 binding sites of Aluminium where determined in the Human ATL1 Mutant - F151S Bound to GDPALF4-, PDB code: 6b9f:
Jump to Aluminium binding site number: 1; 2;
Aluminium binding site 1 out of 2 in 6b9f
Go back to
Aluminium binding site 1 out
of 2 in the Human ATL1 Mutant - F151S Bound to GDPALF4-
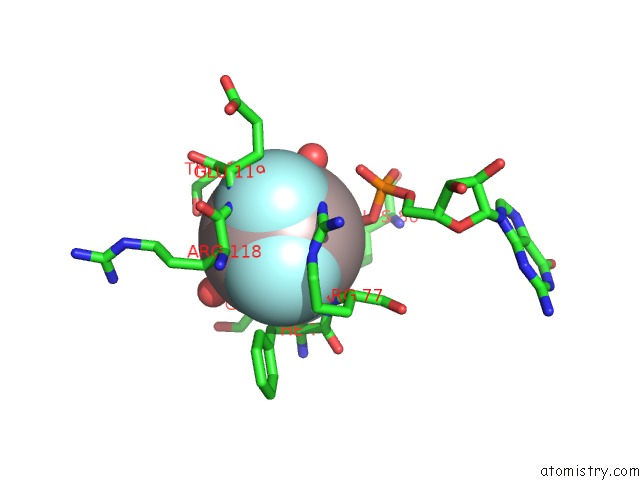
Mono view
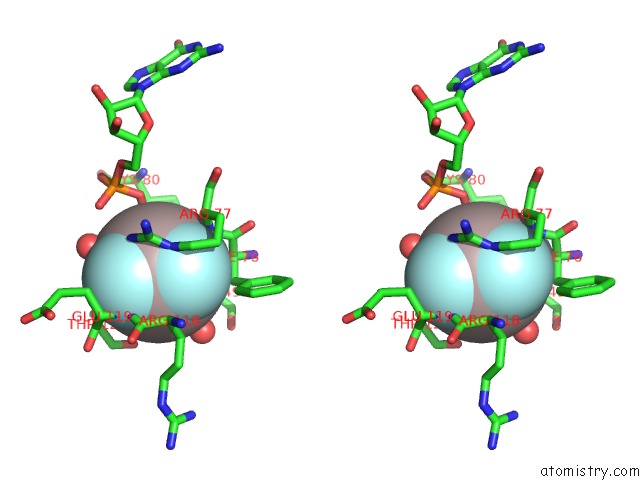
Stereo pair view

Mono view

Stereo pair view
A full contact list of Aluminium with other atoms in the Al binding
site number 1 of Human ATL1 Mutant - F151S Bound to GDPALF4- within 5.0Å range:
|
Aluminium binding site 2 out of 2 in 6b9f
Go back to
Aluminium binding site 2 out
of 2 in the Human ATL1 Mutant - F151S Bound to GDPALF4-
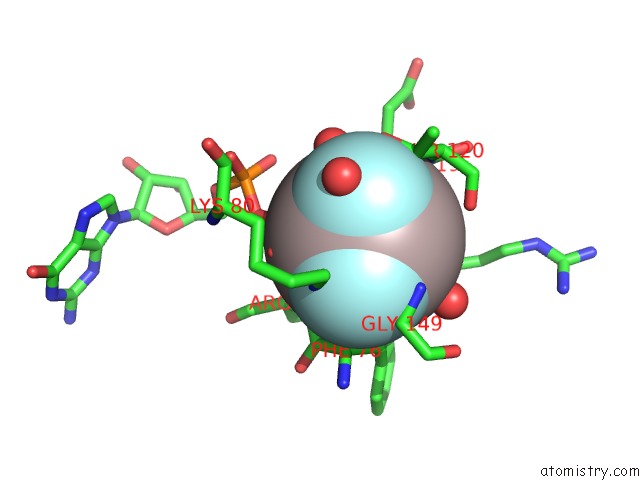
Mono view
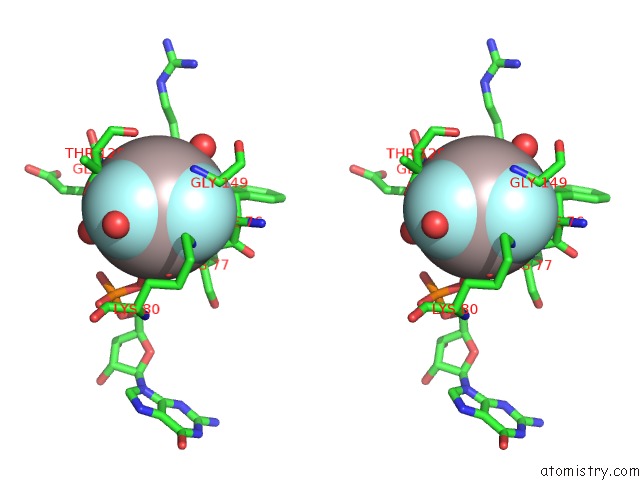
Stereo pair view

Mono view

Stereo pair view
A full contact list of Aluminium with other atoms in the Al binding
site number 2 of Human ATL1 Mutant - F151S Bound to GDPALF4- within 5.0Å range:
|
Reference:
J.P.O'donnell,
L.J.Byrnes,
R.B.Cooley,
H.Sondermann.
A Hereditary Spastic Paraplegia-Associated Atlastin Variant Exhibits Defective Allosteric Coupling in the Catalytic Core. J. Biol. Chem. V. 293 687 2018.
ISSN: ESSN 1083-351X
PubMed: 29180453
DOI: 10.1074/JBC.RA117.000380
Page generated: Sun Jul 6 21:58:27 2025
ISSN: ESSN 1083-351X
PubMed: 29180453
DOI: 10.1074/JBC.RA117.000380
Last articles
Ca in 6DMNCa in 6DMS
Ca in 6DMY
Ca in 6DLH
Ca in 6DIC
Ca in 6DHZ
Ca in 6DIA
Ca in 6DHT
Ca in 6DHY
Ca in 6DGM