Aluminium »
PDB 6i4i-7jl2 »
6k7m »
Aluminium in PDB 6k7m: Cryo-Em Structure of the Human P4-Type Flippase ATP8A1-CDC50 (E2PI-Pl State)
Enzymatic activity of Cryo-Em Structure of the Human P4-Type Flippase ATP8A1-CDC50 (E2PI-Pl State)
All present enzymatic activity of Cryo-Em Structure of the Human P4-Type Flippase ATP8A1-CDC50 (E2PI-Pl State):
7.6.2.1;
7.6.2.1;
Other elements in 6k7m:
The structure of Cryo-Em Structure of the Human P4-Type Flippase ATP8A1-CDC50 (E2PI-Pl State) also contains other interesting chemical elements:
| Fluorine | (F) | 4 atoms |
| Magnesium | (Mg) | 1 atom |
Aluminium Binding Sites:
The binding sites of Aluminium atom in the Cryo-Em Structure of the Human P4-Type Flippase ATP8A1-CDC50 (E2PI-Pl State)
(pdb code 6k7m). This binding sites where shown within
5.0 Angstroms radius around Aluminium atom.
In total only one binding site of Aluminium was determined in the Cryo-Em Structure of the Human P4-Type Flippase ATP8A1-CDC50 (E2PI-Pl State), PDB code: 6k7m:
In total only one binding site of Aluminium was determined in the Cryo-Em Structure of the Human P4-Type Flippase ATP8A1-CDC50 (E2PI-Pl State), PDB code: 6k7m:
Aluminium binding site 1 out of 1 in 6k7m
Go back to
Aluminium binding site 1 out
of 1 in the Cryo-Em Structure of the Human P4-Type Flippase ATP8A1-CDC50 (E2PI-Pl State)
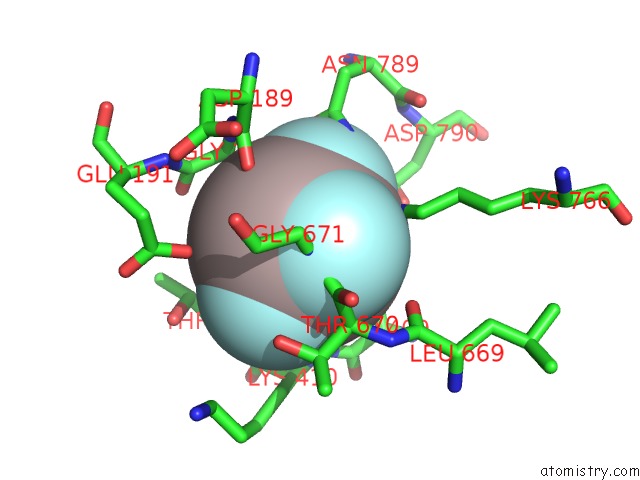
Mono view
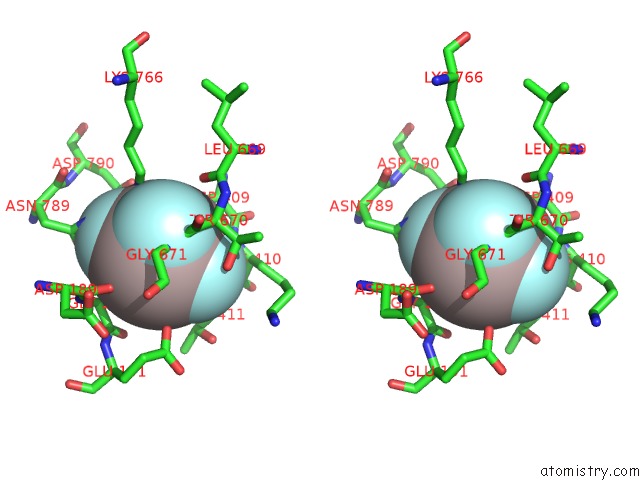
Stereo pair view

Mono view

Stereo pair view
A full contact list of Aluminium with other atoms in the Al binding
site number 1 of Cryo-Em Structure of the Human P4-Type Flippase ATP8A1-CDC50 (E2PI-Pl State) within 5.0Å range:
|
Reference:
M.Hiraizumi,
K.Yamashita,
T.Nishizawa,
O.Nureki.
Cryo-Em Structures Capture the Transport Cycle of the P4-Atpase Flippase. Science V. 365 1149 2019.
ISSN: ESSN 1095-9203
PubMed: 31416931
DOI: 10.1126/SCIENCE.AAY3353
Page generated: Sun Jul 6 21:59:48 2025
ISSN: ESSN 1095-9203
PubMed: 31416931
DOI: 10.1126/SCIENCE.AAY3353
Last articles
Ca in 6D5DCa in 6D51
Ca in 6D3W
Ca in 6D50
Ca in 6D3U
Ca in 6D4K
Ca in 6D2C
Ca in 6D3B
Ca in 6D2S
Ca in 6D05