Aluminium »
PDB 7jl3-7t22 »
7kue »
Aluminium in PDB 7kue: Cryoem Structure of Yeast Tfiik (KIN28/CCL1/TFB3) Complex
Enzymatic activity of Cryoem Structure of Yeast Tfiik (KIN28/CCL1/TFB3) Complex
All present enzymatic activity of Cryoem Structure of Yeast Tfiik (KIN28/CCL1/TFB3) Complex:
2.7.11.23;
2.7.11.23;
Other elements in 7kue:
The structure of Cryoem Structure of Yeast Tfiik (KIN28/CCL1/TFB3) Complex also contains other interesting chemical elements:
| Fluorine | (F) | 3 atoms |
Aluminium Binding Sites:
The binding sites of Aluminium atom in the Cryoem Structure of Yeast Tfiik (KIN28/CCL1/TFB3) Complex
(pdb code 7kue). This binding sites where shown within
5.0 Angstroms radius around Aluminium atom.
In total only one binding site of Aluminium was determined in the Cryoem Structure of Yeast Tfiik (KIN28/CCL1/TFB3) Complex, PDB code: 7kue:
In total only one binding site of Aluminium was determined in the Cryoem Structure of Yeast Tfiik (KIN28/CCL1/TFB3) Complex, PDB code: 7kue:
Aluminium binding site 1 out of 1 in 7kue
Go back to
Aluminium binding site 1 out
of 1 in the Cryoem Structure of Yeast Tfiik (KIN28/CCL1/TFB3) Complex
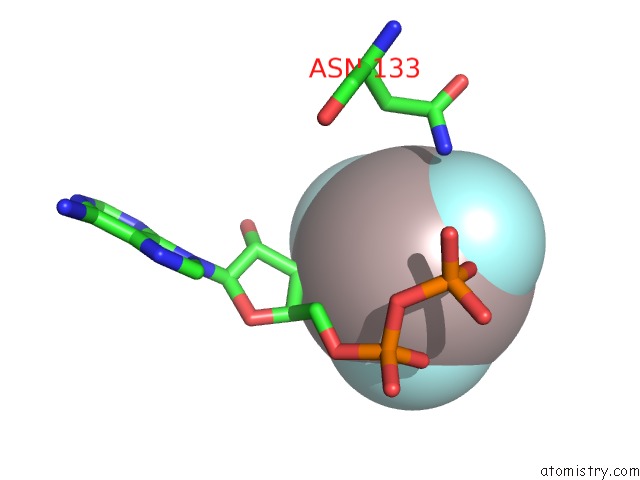
Mono view
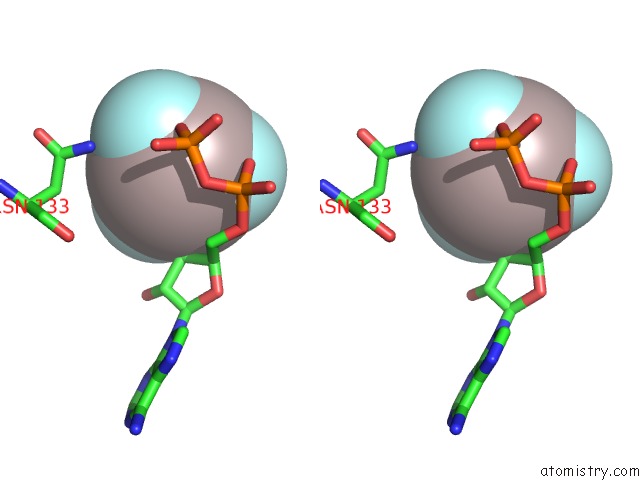
Stereo pair view

Mono view

Stereo pair view
A full contact list of Aluminium with other atoms in the Al binding
site number 1 of Cryoem Structure of Yeast Tfiik (KIN28/CCL1/TFB3) Complex within 5.0Å range:
|
Reference:
T.Van Eeuwen,
T.Li,
H.J.Kim,
J.J.Gorbea Colon,
M.I.Parker,
R.L.Dunbrack,
B.A.Garcia,
K.L.Tsai,
K.Murakami.
Structure of Tfiik For Phosphorylation of Ctd of Rna Polymerase II. Sci Adv V. 7 2021.
ISSN: ESSN 2375-2548
PubMed: 33827808
DOI: 10.1126/SCIADV.ABD4420
Page generated: Sun Jul 6 22:04:08 2025
ISSN: ESSN 2375-2548
PubMed: 33827808
DOI: 10.1126/SCIADV.ABD4420
Last articles
F in 4JTWF in 4JSX
F in 4JSV
F in 4JTQ
F in 4JSC
F in 4JSJ
F in 4JSM
F in 4JQG
F in 4JSI
F in 4JQ2