Aluminium »
PDB 6i4i-7jl2 »
7jl0 »
Aluminium in PDB 7jl0: Cryo-Em Structure of MDA5-Dsrna in Complex with TRIM65 Pspry Domain (Monomer)
Enzymatic activity of Cryo-Em Structure of MDA5-Dsrna in Complex with TRIM65 Pspry Domain (Monomer)
All present enzymatic activity of Cryo-Em Structure of MDA5-Dsrna in Complex with TRIM65 Pspry Domain (Monomer):
3.6.4.13;
3.6.4.13;
Other elements in 7jl0:
The structure of Cryo-Em Structure of MDA5-Dsrna in Complex with TRIM65 Pspry Domain (Monomer) also contains other interesting chemical elements:
| Fluorine | (F) | 4 atoms |
| Magnesium | (Mg) | 1 atom |
| Zinc | (Zn) | 1 atom |
Aluminium Binding Sites:
The binding sites of Aluminium atom in the Cryo-Em Structure of MDA5-Dsrna in Complex with TRIM65 Pspry Domain (Monomer)
(pdb code 7jl0). This binding sites where shown within
5.0 Angstroms radius around Aluminium atom.
In total only one binding site of Aluminium was determined in the Cryo-Em Structure of MDA5-Dsrna in Complex with TRIM65 Pspry Domain (Monomer), PDB code: 7jl0:
In total only one binding site of Aluminium was determined in the Cryo-Em Structure of MDA5-Dsrna in Complex with TRIM65 Pspry Domain (Monomer), PDB code: 7jl0:
Aluminium binding site 1 out of 1 in 7jl0
Go back to
Aluminium binding site 1 out
of 1 in the Cryo-Em Structure of MDA5-Dsrna in Complex with TRIM65 Pspry Domain (Monomer)
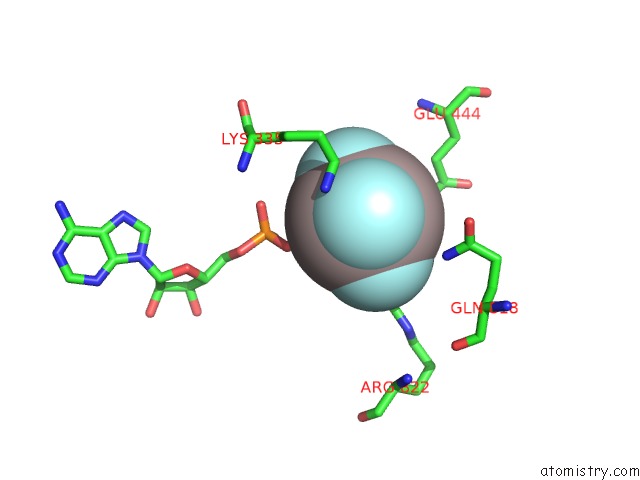
Mono view
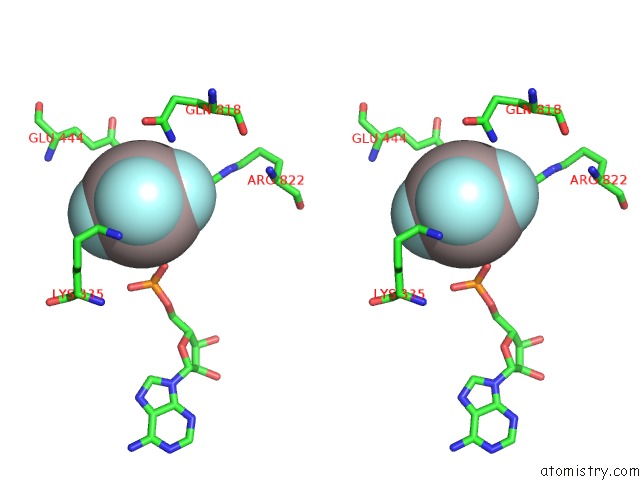
Stereo pair view

Mono view

Stereo pair view
A full contact list of Aluminium with other atoms in the Al binding
site number 1 of Cryo-Em Structure of MDA5-Dsrna in Complex with TRIM65 Pspry Domain (Monomer) within 5.0Å range:
|
Reference:
K.Kato,
S.Ahmad,
Z.Zhu,
J.M.Young,
X.Mu,
S.Park,
H.S.Malik,
S.Hur.
Structural Analysis of Rig-I-Like Receptors Reveals Ancient Rules of Engagement Between Diverse Rna Helicases and Trim Ubiquitin Ligases Mol.Cell 2020.
ISSN: ISSN 1097-2765
Page generated: Sun Jul 6 22:03:42 2025
ISSN: ISSN 1097-2765
Last articles
Mn in 9LJUMn in 9LJW
Mn in 9LJS
Mn in 9LJR
Mn in 9LJT
Mn in 9LJV
Mg in 9UA2
Mg in 9R96
Mg in 9VM1
Mg in 9P01