Aluminium »
PDB 7t3b-8ien »
8eew »
Aluminium in PDB 8eew: Cryoem of the Soluble OPA1 Dimer From the Gdp-Alfx Bound Helical Assembly on A Lipid Membrane
Other elements in 8eew:
The structure of Cryoem of the Soluble OPA1 Dimer From the Gdp-Alfx Bound Helical Assembly on A Lipid Membrane also contains other interesting chemical elements:
| Fluorine | (F) | 8 atoms |
| Potassium | (K) | 2 atoms |
| Magnesium | (Mg) | 2 atoms |
Aluminium Binding Sites:
The binding sites of Aluminium atom in the Cryoem of the Soluble OPA1 Dimer From the Gdp-Alfx Bound Helical Assembly on A Lipid Membrane
(pdb code 8eew). This binding sites where shown within
5.0 Angstroms radius around Aluminium atom.
In total 2 binding sites of Aluminium where determined in the Cryoem of the Soluble OPA1 Dimer From the Gdp-Alfx Bound Helical Assembly on A Lipid Membrane, PDB code: 8eew:
Jump to Aluminium binding site number: 1; 2;
In total 2 binding sites of Aluminium where determined in the Cryoem of the Soluble OPA1 Dimer From the Gdp-Alfx Bound Helical Assembly on A Lipid Membrane, PDB code: 8eew:
Jump to Aluminium binding site number: 1; 2;
Aluminium binding site 1 out of 2 in 8eew
Go back to
Aluminium binding site 1 out
of 2 in the Cryoem of the Soluble OPA1 Dimer From the Gdp-Alfx Bound Helical Assembly on A Lipid Membrane
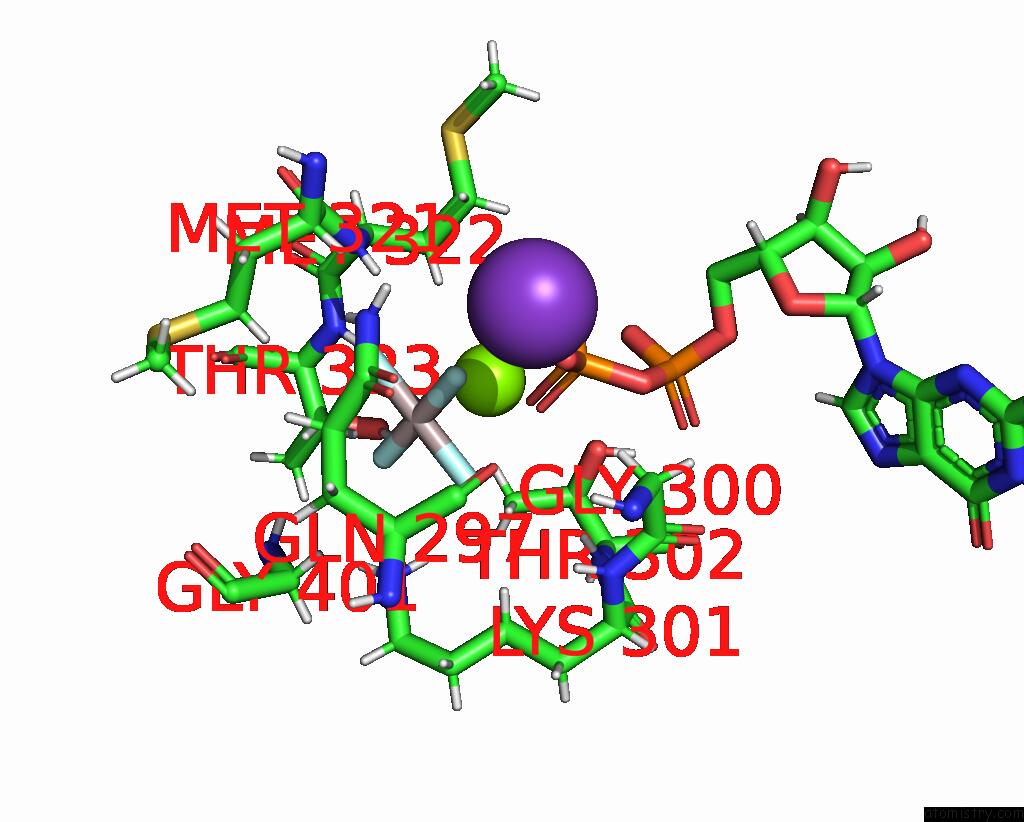
Mono view
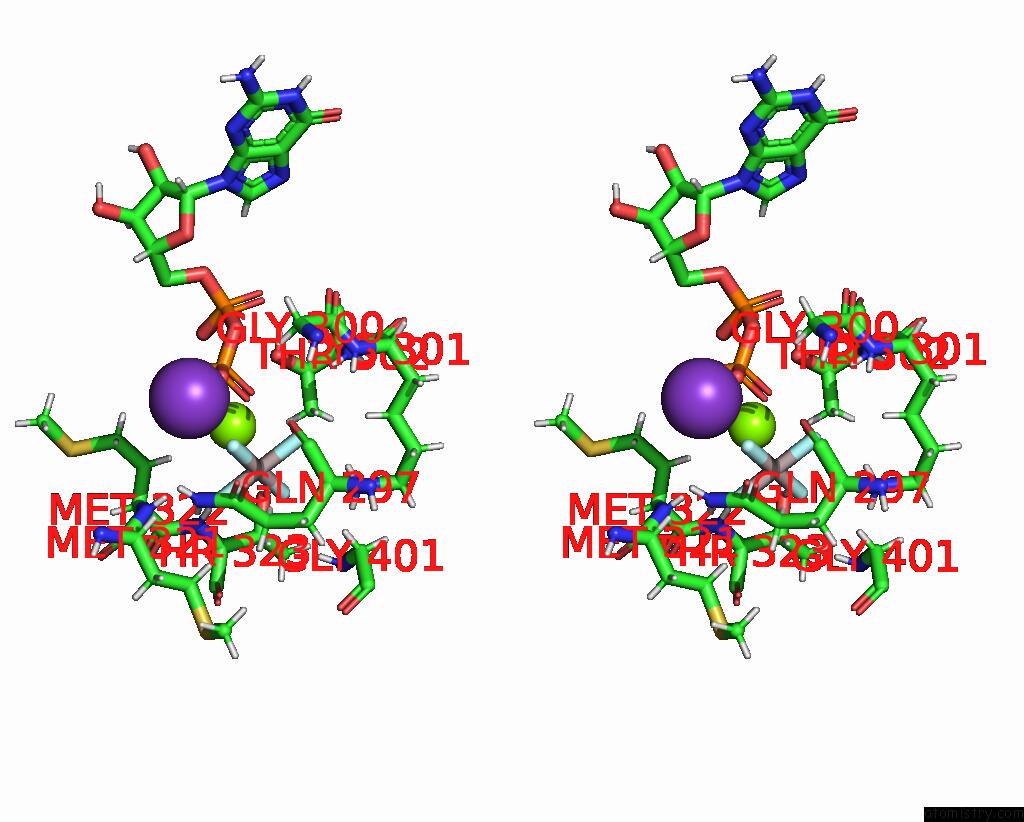
Stereo pair view

Mono view

Stereo pair view
A full contact list of Aluminium with other atoms in the Al binding
site number 1 of Cryoem of the Soluble OPA1 Dimer From the Gdp-Alfx Bound Helical Assembly on A Lipid Membrane within 5.0Å range:
|
Aluminium binding site 2 out of 2 in 8eew
Go back to
Aluminium binding site 2 out
of 2 in the Cryoem of the Soluble OPA1 Dimer From the Gdp-Alfx Bound Helical Assembly on A Lipid Membrane
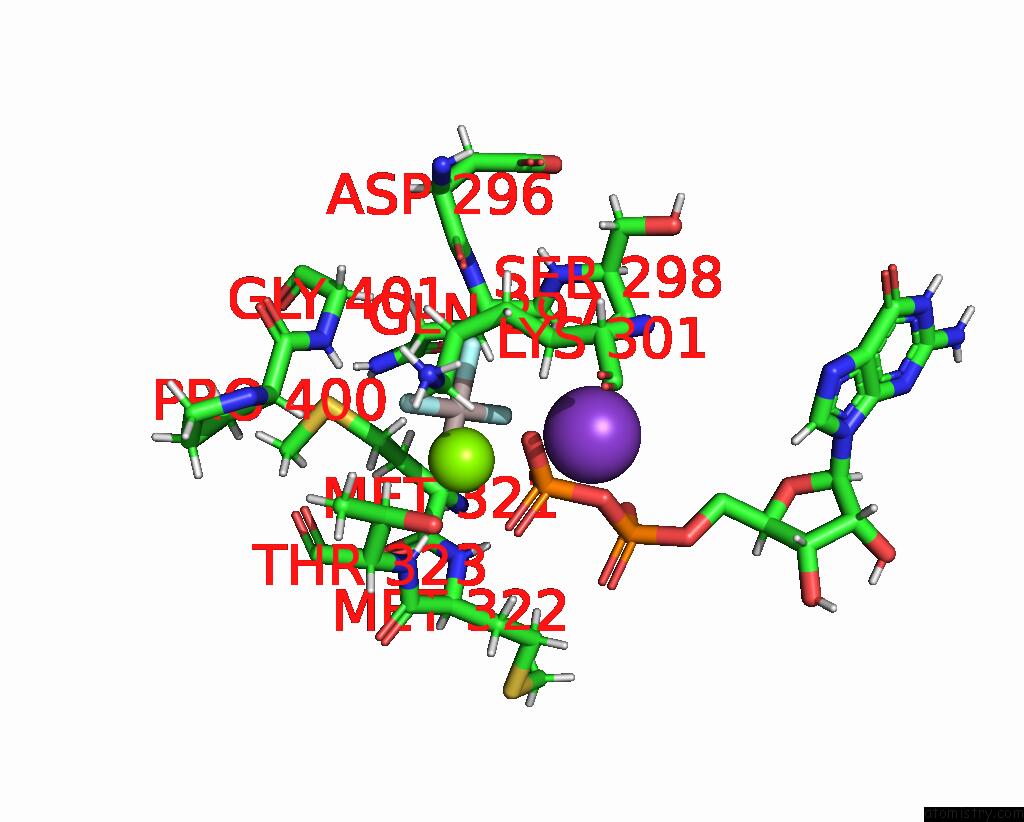
Mono view
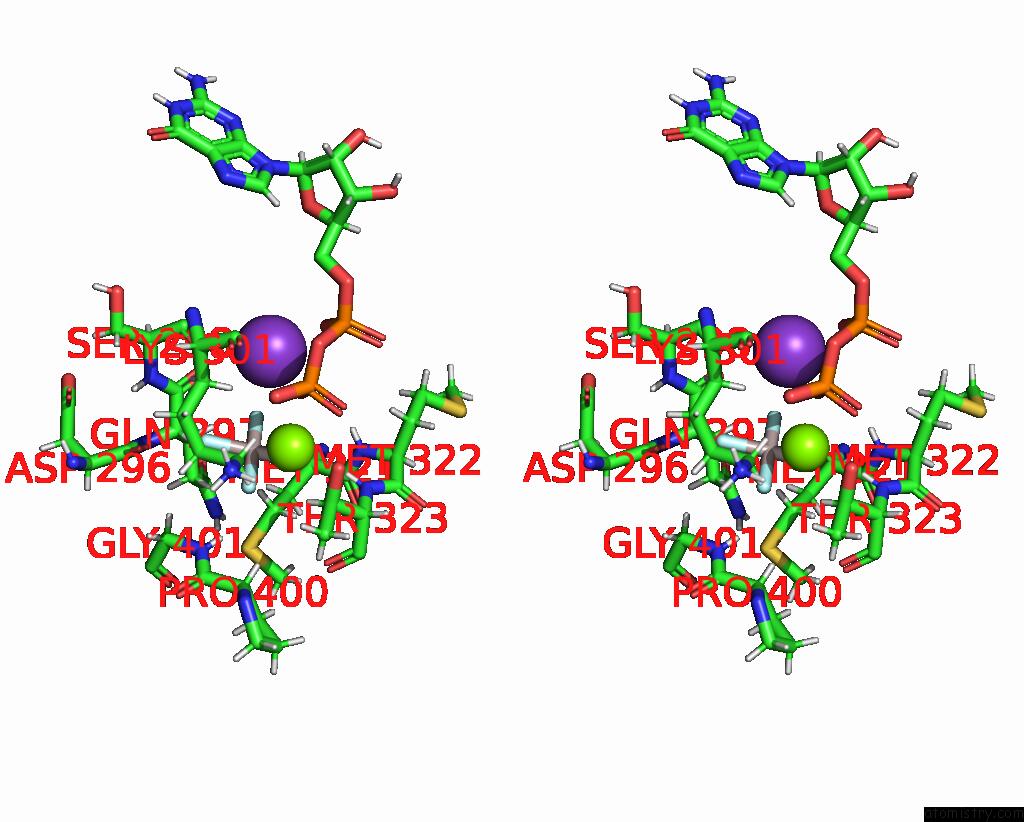
Stereo pair view

Mono view

Stereo pair view
A full contact list of Aluminium with other atoms in the Al binding
site number 2 of Cryoem of the Soluble OPA1 Dimer From the Gdp-Alfx Bound Helical Assembly on A Lipid Membrane within 5.0Å range:
|
Reference:
S.B.Nyenhuis,
X.Wu,
A.E.Stanton,
M.P.Strub,
Y.I.Yim,
B.Canagarajah,
J.E.Hinshaw.
OPA1 Helical Structures Give Perspective to Mitochondrial Dysfunction Nature 2023.
ISSN: ESSN 1476-4687
Page generated: Sun Jul 6 22:18:47 2025
ISSN: ESSN 1476-4687
Last articles
Mn in 3WVIMn in 3WVH
Mn in 3WU2
Mn in 3WQO
Mn in 3WKJ
Mn in 3WOG
Mn in 3WRO
Mn in 3WCS
Mn in 3WEI
Mn in 3WI8