Aluminium »
PDB 8uqo-9q97 »
9f2l »
Aluminium in PDB 9f2l: Cryo-Em Structure of the I923V MDA5-Dsrna Filament with Adp-ALF4 Bound and 73-Degree Helical Twist
Enzymatic activity of Cryo-Em Structure of the I923V MDA5-Dsrna Filament with Adp-ALF4 Bound and 73-Degree Helical Twist
All present enzymatic activity of Cryo-Em Structure of the I923V MDA5-Dsrna Filament with Adp-ALF4 Bound and 73-Degree Helical Twist:
3.6.4.13;
3.6.4.13;
Other elements in 9f2l:
The structure of Cryo-Em Structure of the I923V MDA5-Dsrna Filament with Adp-ALF4 Bound and 73-Degree Helical Twist also contains other interesting chemical elements:
| Fluorine | (F) | 4 atoms |
| Zinc | (Zn) | 1 atom |
Aluminium Binding Sites:
The binding sites of Aluminium atom in the Cryo-Em Structure of the I923V MDA5-Dsrna Filament with Adp-ALF4 Bound and 73-Degree Helical Twist
(pdb code 9f2l). This binding sites where shown within
5.0 Angstroms radius around Aluminium atom.
In total only one binding site of Aluminium was determined in the Cryo-Em Structure of the I923V MDA5-Dsrna Filament with Adp-ALF4 Bound and 73-Degree Helical Twist, PDB code: 9f2l:
In total only one binding site of Aluminium was determined in the Cryo-Em Structure of the I923V MDA5-Dsrna Filament with Adp-ALF4 Bound and 73-Degree Helical Twist, PDB code: 9f2l:
Aluminium binding site 1 out of 1 in 9f2l
Go back to
Aluminium binding site 1 out
of 1 in the Cryo-Em Structure of the I923V MDA5-Dsrna Filament with Adp-ALF4 Bound and 73-Degree Helical Twist
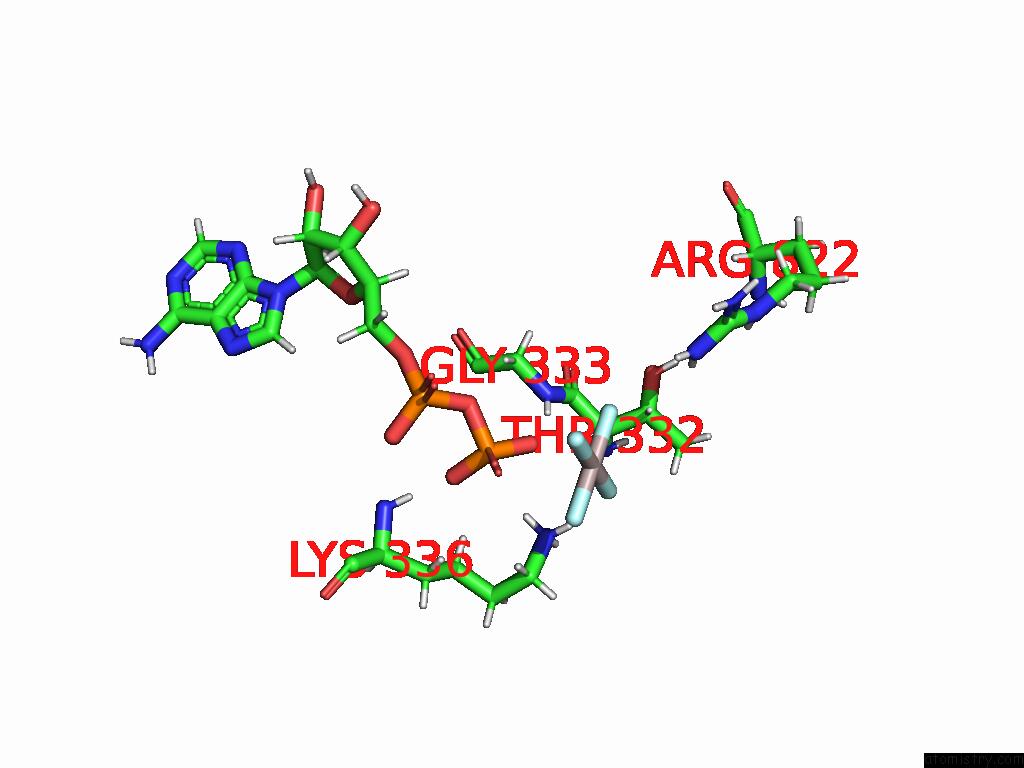
Mono view
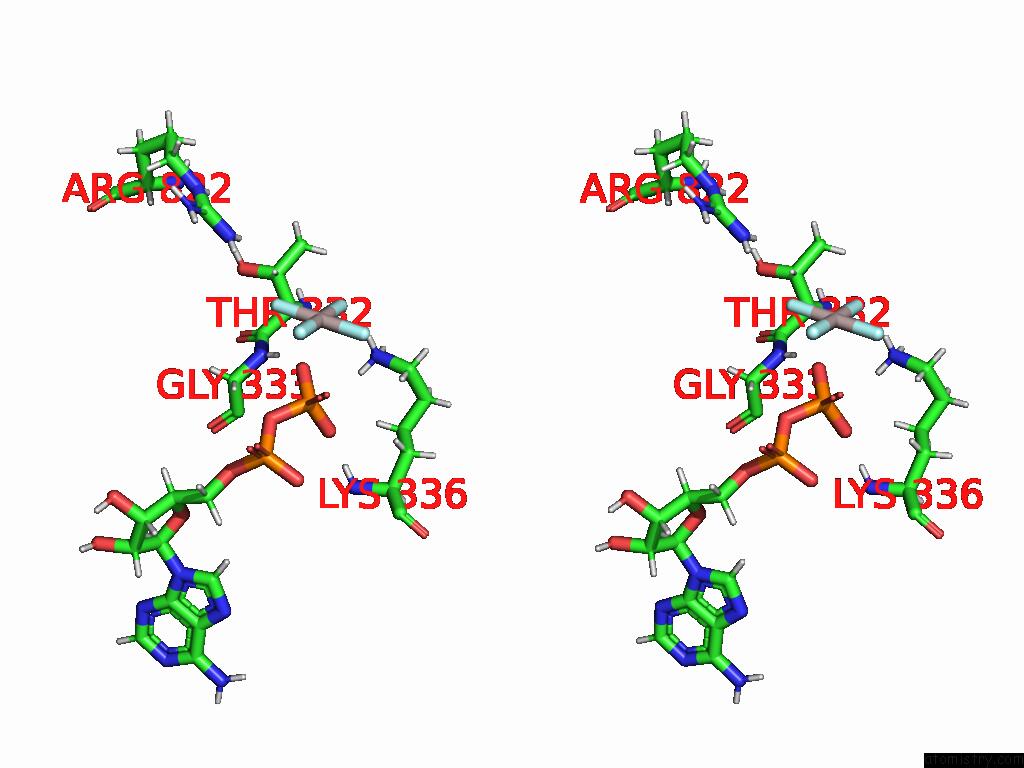
Stereo pair view

Mono view

Stereo pair view
A full contact list of Aluminium with other atoms in the Al binding
site number 1 of Cryo-Em Structure of the I923V MDA5-Dsrna Filament with Adp-ALF4 Bound and 73-Degree Helical Twist within 5.0Å range:
|
Reference:
R.Singh,
A.Herrero Del Valle,
J.D.Joiner,
M.Zwaagstra,
B.J.Ferguson,
F.J.M.Van Kuppeveld,
Y.Modis.
Molecular Basis of Autoimmune Disease Protection By MDA5 Variants Biorxiv 2024.
ISSN: ISSN 2692-8205
DOI: 10.1101/2024.10.03.616466
Page generated: Sun Jul 6 22:41:48 2025
ISSN: ISSN 2692-8205
DOI: 10.1101/2024.10.03.616466
Last articles
K in 9NESK in 9PHG
K in 9NEI
K in 9NED
K in 9NEC
K in 9NEG
K in 9CWU
K in 9CVB
K in 9CVA
K in 9COM